Microbial Interface Biology

 The Research Group Microbial Interface Biology focuses on developing innovative therapeutic approaches targeting both the host and the pathogen to combat tuberculosis as a global health threat. The research group concentrates on two complementary areas: 1) deciphering host cell processes relevant to M. tuberculosis infections, with a focus on macrophages as a central cellular niche, and 2) identifying and evaluating novel compounds with anti-mycobacterial activity. Both topics are addressed using a broad spectrum of cellular, biochemical and microbiological methods. The research group has many years of comprehensive expertise in the isolation and infection of primary human macrophages as well as in the biology and genetic modification of mycobacteria.
The Research Group Microbial Interface Biology focuses on developing innovative therapeutic approaches targeting both the host and the pathogen to combat tuberculosis as a global health threat. The research group concentrates on two complementary areas: 1) deciphering host cell processes relevant to M. tuberculosis infections, with a focus on macrophages as a central cellular niche, and 2) identifying and evaluating novel compounds with anti-mycobacterial activity. Both topics are addressed using a broad spectrum of cellular, biochemical and microbiological methods. The research group has many years of comprehensive expertise in the isolation and infection of primary human macrophages as well as in the biology and genetic modification of mycobacteria.
Our detailed studies in the field of host pathogen interaction have identified previously unrecognized host pathways associated with the regulation of lipid metabolism in the context of mycobacterial infection. Thus, we currently aim to develop an innovative, host-directed therapeutic approach through the use of novel, highly specific small molecule inhibitors that target such pathways, thereby limiting the access of M. tuberculosis to fatty acids – its preferred carbon source in vivo. The studies involve the use of new fluorescent reporter strains, transcriptional knockdowns in M. tuberculosis and cellular model systems with a focus on lipid droplet dynamics and their interaction with the TB causing pathogen.
In addition, the FG has a long-standing interest in early-stage antitubercular drug discovery, and is an integral part of the preclinical drug development pipelines at both the FZB and within the TTU Tuberculosis at the German Centre for Infection Research (DZIF). The group’s expertise is also documented in its active collaboration with over 20 national and international partner institutions in various TB drug discovery projects. The expansion of in vitro drug testing capacity and capability is being constantly pursued within the group, including the ongoing development of genetically modified M. tuberculosis strains for more efficient characterization of new drugs and their targets. Together, these approaches emphasize our position as a key European hub in antitubercular in vitro drug profiling and the early phases of TB drug development. Overall, the group successfully combines fundamental scientific questions with a clear translational perspective.
I. The WNT signaling pathway in M. tuberculosis infection – from novel mediators to host-directed therapy
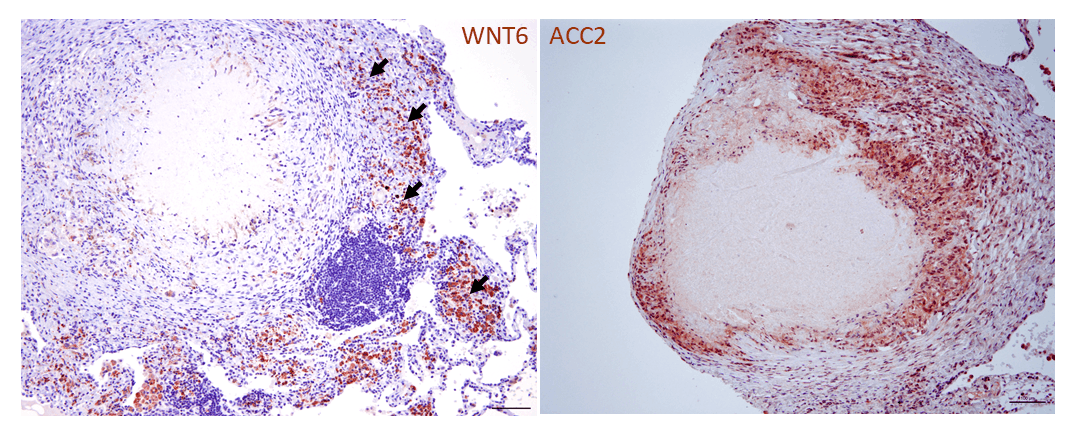
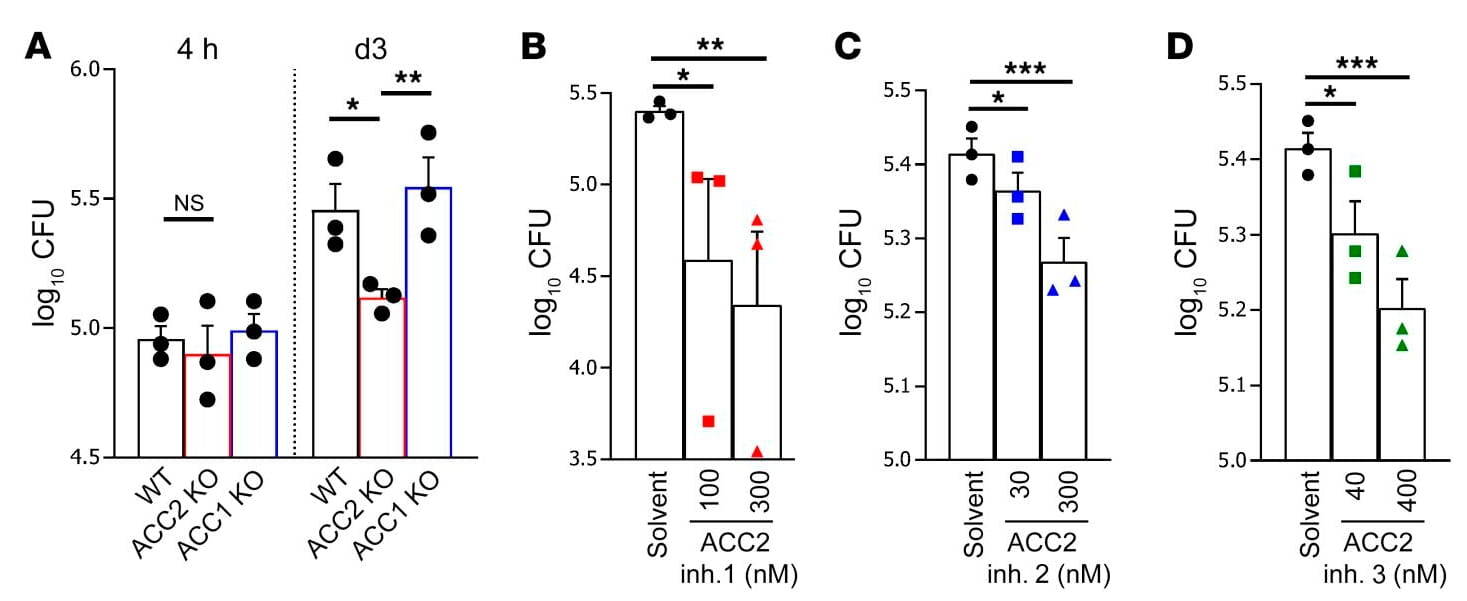
Building on our previous work on the functional significance of pattern recognition receptors in M. tuberculosis infection, systematic analyses of mycobacterial-infected macrophages have for the first time demonstrated regulatory functions for components of the evolutionarily highly conserved Wingless/Integrase 1 (WNT) signaling pathway in pulmonary tuberculosis. WNT proteins have pro-inflammatory or anti-inflammatory effects on macrophages and other cells of the immune system and sometimes also influence the development of bacterial numbers during infection. WNT6 plays a special role in this context, and we were able to demonstrate its prominent expression both in granulomatous lesions in the lungs of M. tuberculosis-infected mice and in the lung tissue of TB patients (Fig. 1a; Brandenburg et al., J. Clin Invest 2021). Under our leadership and in close collaboration with eight other research groups at the FZB, WNT6 was identified through comprehensive functional analyses as a key regulator of foam macrophage formation. These lipid-rich foam cells form a central intracellular habitat for M. tuberculosis during tuberculosis infection. By modulating WNT6-induced signal transduction, specifically through targeted inhibition of acetyl-CoA carboxylase 2 using both genetic and pharmacological methods (Fig. 1b), we were able to significantly inhibit the growth of M. tuberculosis, both in vitro and in vivo (Brandenburg et al., J. Clin Invest 2021). This targeted modulation of neutral lipid metabolism identified by us represents a promising prospect for a new host-specific adjunctive therapy for TB. (Patent granted: Reiling N & Brandenburg J; EP3481392B1). This topic is currently being investigated in more depth in further studies on lipid metabolism during M. tuberculosis infection, involving mouse strains with varying degrees of susceptibility.
Selected project references:
- Brandenburg J, et al.; J Clin Invest. 2021 Aug 16;131(16):e141833. doi: 10.1172/JCI141833.
- Reiling N & Brandenburg J; EP3481392B1 (patent granted).
- Brandenburg J, et al.; ACS Infect Dis. 2022 Jul 8;8(7):1303-1315. doi: 10.1021/acsinfecdis.2c00075.
- Brandenburg J & Reiling N.; Front Immunol. 2016 Dec 26;7:635. doi: 10.3389/fimmu.2016.00635.
- Schaale K, et al.; J Immunol. 2013 Nov 15;191(10):5182-95. doi: 10.4049/jimmunol.1201819.
- Schaale K, et al.; Eur J Cell Biol. 2011 Jun-Jul;90(6-7):553-9. doi: 10.1016/j.ejcb.2010.11.004.
- Neumann J, et al.; FASEB J. 2010 Nov;24(11):4599-612. doi: 10.1096/fj.10-160994.
- Blumenthal A, et al.; Blood. 2006 Aug 1;108(3):965-73. doi: 10.1182/blood-2005-12-5046.
II. Phosphatidylinositols containing tuberculostaric acid as markers for bacterial load in tuberculosis
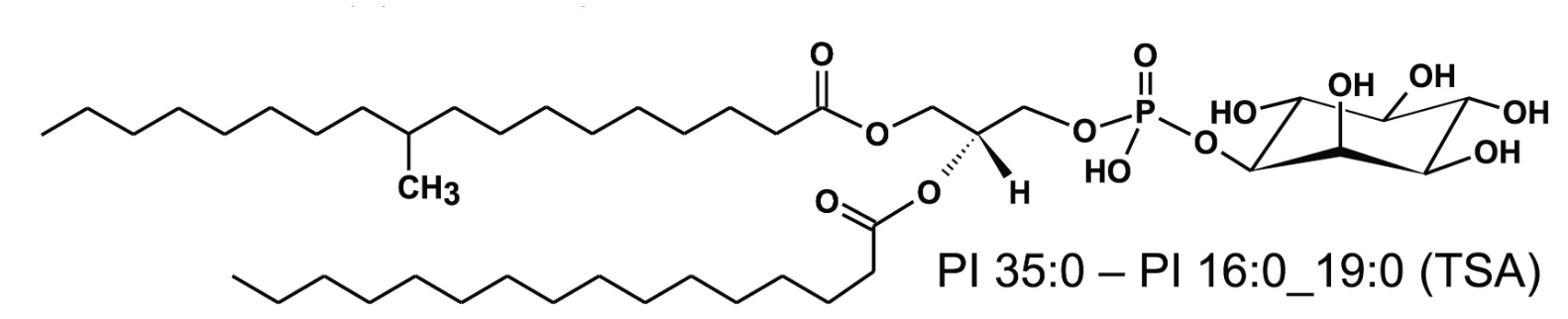
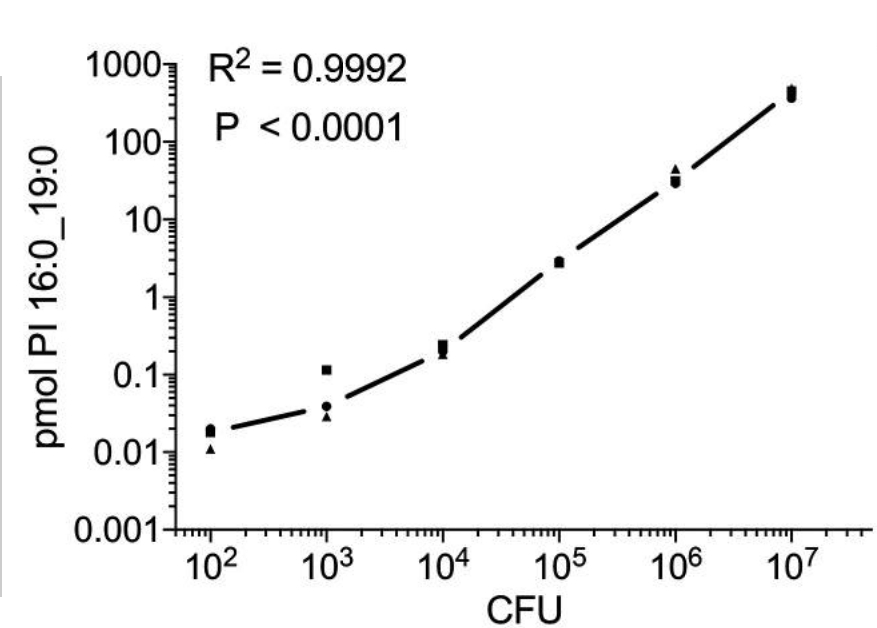
Also of translational importance – but here relating to the diagnosis of TB – is the detection of specific M. tuberculosis lipids, which can be used as biomarkers for monitoring tuberculosis therapy (Brandenburg et al., ACS Infect. Dis. 2022). Using lipidomic approaches in close cooperation with the Research Group Bioanalytical Chemistry, we were able to show that phosphatidylinositols (PIs) containing tuberculostearic acid (TSA) are molecular markers for infection with clinically relevant M. tuberculosis complex strains and indicate bacterial load. For the most common lipid marker, detection limits of ∼102 and ∼103 colony-forming units (CFU) were determined for bacterial and cell culture systems, respectively (Fig. 2). A complete analytical method, including sample preparation, was developed that can be performed within one day – about 30 times faster than conventional methods based on bacterial cultures. Using this indirect and culture-free detection method, we were able to determine the pathogen load in infected murine macrophages, human neutrophils and murine lung tissue. These marker lipids derived from mycobacterial PIs were found in higher concentrations in the mononuclear cells of the peripheral blood of TB patients than in healthy individuals. Furthermore, in a small cohort of antibiotic-responsive TB patients, elevated concentrations of these molecular markers were detected at the start of therapy, which decreased after successful anti-TB treatment. Thus, the concentration of TSA-containing PIs can be used as a correlate for mycobacterial load in experimental models and in vitro systems and could also represent a clinically relevant tool for monitoring the severity of TB in infected patients.
Selected project references:
- Brandenburg J, et al. J Clin Invest. 2021 Aug 16;131(16):e141833. doi: 10.1172/JCI141833.
- Brandenburg J, et al.; ACS Infect Dis. 2022 Jul 8;8(7):1303-1315. doi: 10.1021/acsinfecdis.2c00075.
III. Multiple Strategies of M. tuberculosis for surviving Spermine-induced Stress
The discovery of molecules that could shorten the treatment period of tuberculosis (TB) holds the promise of improving the current regimen. Polyamines are organic polycationic alkylamines found in millimolar concentrations in all living cells. Spermine (Spm) is a polyamine that was shown (more than 70 years ago) to be toxic to M. tuberculosis. The actinomycete Streptomyces coelicolor, a close relative of M. tuberculosis, makes use of a gamma-glutamylation pathway to functionally neutralize Spm. We therefore considered whether a similar pathway would be functional in M. tuberculosis and investigated how this bacterium defends itself against the toxic effects of this polyamine since it is produced by host macrophages. We observed that Spm inhibits M. tuberculosis growth and that the enzyme GlnA3Mt (Rv1878) functions as a γ‑glutamylspermine synthetase, preferentially acting on spermine over other polyamines. However, deleting the glnA3 gene did not make the bacteria more sensitive to spermine, and the gene’s expression was unaffected by spermine exposure. Instead, the efflux pump gene mmr (Rv3065) was strongly upregulated, indicating that spermine detoxification can occur through active expulsion rather than enzymatic modification. Further experiments showed distinct roles for other efflux pumps and enzymes in spermine resistance and stress responses. The Δrv1877 mutant was sensitive to spermine stress, implicating Rv1877 (a multidrug MFS-type efflux pump) in spermine export. In contrast, Δrv0191 was insensitive to spermine but sensitive to oxidative stress, implicating role in transporting low‑molecular‑weight thiols. Meanwhile, Δrv1878 (GlnA3-deficient) exhibited enhanced growth under iron starvation but greater sensitivity to cell wall stress, implying that Rv1878 may contribute to iron regulation and cell wall maintenance. Overall, our studies suggest that M. tuberculosis uses multiple, specialized mechanisms to survive spermine toxicity and related cellular stresses.
Selected project references:
- C. Sao Emani & N. Reiling; Microbiol Spectr. 2024 Jan 11;12(1):e0356823. doi: 10.1128/spectrum.03568-23.
- C. Sao Emani & N. Reiling; Front Microbiol. 2024 Mar 4;15:1359188. doi: 0.3389/fmicb.2024.1359188.
- Krysenko S, et al. J Bacteriol. 2025 Feb 20;207(2):e0043924. doi: 10.1128/jb.00439-24.
IV. ‘New drugs wanted’ – Fast and relevant test systems for identifying new active substances against M. tuberculosis
In 2024, tuberculosis regained its position as the leading cause of death from a single infectious agent, and currently sits among the ten leading causes of death worldwide. Thus, there is an urgent need for new antibiotics to improve the treatment outcomes of tuberculosis patients, especially those infected with strains resistant to the best currently available front-line drugs. Against this backdrop, we have established various in vitro test systems in recent years to quickly and reliably identify and characterize potential new anti-TB molecules. From 384-well plate fluorescence-mediated compound library screening and dose-response growth-inhibition assays (IC50 determination), to time kill assays, intramacrophage activity, resistant mutant generation and bespoke target-identification/validation methods, our comprehensive early-stage antitubercular compound identification and characterization pipeline bridges a crucial gap between hit identification and lead development within the German and European tuberculosis drug development community.
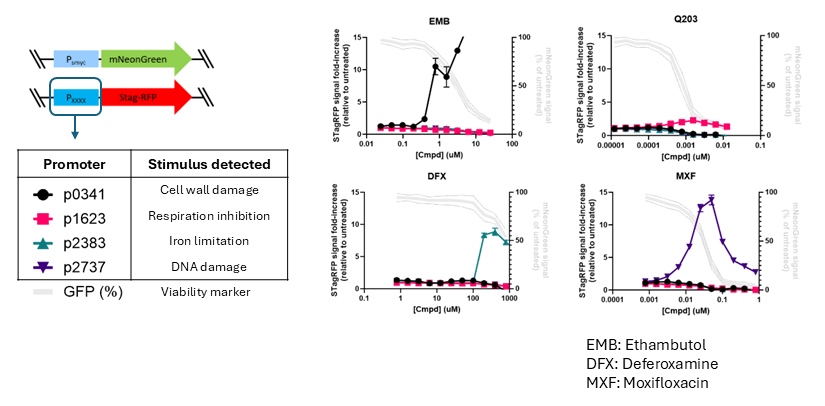
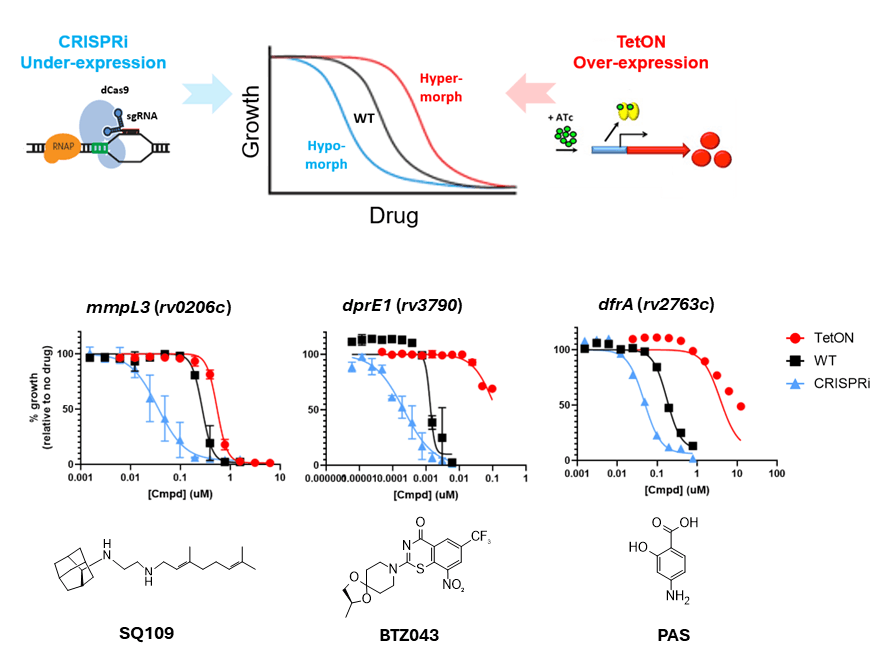
A major focus area within our drug development work is the design and implementation of tools for accelerating compound mechanism of action (MoA)/target elucidation – often a developmental bottleneck in the early stages of phenotypic drug discovery. Determining a compound’s MoA can both facilitate future lead-optimization, for example via structure-guided drug design, as well as help deprioritize compounds with undesirable targets as early as possible, thereby conserving resources for promising leads. In this way, we are harnessing the power and versatility of genetic modification to generate panels of recombinant M. tuberculosis strains that can greatly accelerate compound MoA/target identification. In one approach, we create fluorescent ‘reporter strains’ that via a transcriptional fusion of a stimuli-specific responsive promoter to a fluorescent protein gene allow rapid (within 2-3 days) assessment of specific stress responses induced by compounds of interest (see Fig. 3). In a second approach, we use inducible targeted gene up- and down-regulation for assessing whole cell drug-target engagement, based on the concept of differential drug sensitivity mediated by differential intracellular drug target concentrations (see Fig. 4). In both cases, we are continually updating our strain collections, in order to interrogate as large a selection of MoAs/targets as possible. If any of the tools/techniques mentioned here are of interest for your own research, or if you have compounds that you would like to profile against M. tuberculosis, then please do not hesitate to contact us. We are always enthusiastic to start new drug discovery collaborations!
We are involved in both targeted and phenotypic screens with various cooperation partners, and have strong links with multiple medicinal and organic chemistry groups throughout Europe and beyond, through whom we gain access to critical starting material for identifying new antitubercular compounds. We are proud to be part of the Thematic Translational Transfer Unit Tuberculosis (TTU-TB) of the German Centre for Infection Research (DZIF), which deals with the topic of ‘New Drugs and Regimen’. In addition, we were partners in the EU-funded Marie Skłodowska-Curie Action ‘MepAnti’ (which is a training network for the development of new anti-infectives) and in several research consortia funded by the BMBF/BMFTR. We have recently set up a high-content imaging system that also allows us to identify active substances that target the host rather than the bacteria, with a view to identifying active substances for future host-targeted therapeutic approaches.
Selected project references:
- Kiefer AF, et al.; JACS Au. 2025 Nov 18;5(12):6060-6071. doi: 10.1021/jacsau.5c00950.
- Palme PR, et al.; J Med Chem. 2025 Dec 11;68(23):25274-25289. doi: 10.1021/acs.jmedchem.5c02284.
- Braun-Cornejo M, et al.; JACS Au. 2025 Feb 21;5(3):1146-1156. doi: 10.1021/jacsau.4c00935.
- Braun-Cornejo M, et al.; ACS Omega. 2024 Aug 29;9(36):38160-38168. doi: 10.1021/acsomega.4c05537.
- Du X, et al.; ChemMedChem. 2023 Sep 1;18(17):e202300279. doi: 10.1002/cmdc.202300279.
- Johannsen S, et al. ChemMedChem. 2023 Jun 1;18(11):e202200590. doi: 10.1002/cmdc.202200590.
- Zhu D, et al.; Chem Sci. 2022 Aug 8;13(36):10686-10698. doi: 10.1039/d2sc02371g.
- Richter A, et al.; ACS Med Chem Lett. 2022 Jul 25;13(8):1302-1310. doi: 10.1021/acsmedchemlett.2c00215. Aryal N, et al.;.J Nat Prod. 2022 Mar 25;85(3):530-539. doi: 10.1021/acs.jnatprod.1c01051.
- Brandenburg J, et al. ;.J Clin Invest. 2021 Aug 16;131(16):e141833.
- Jumde RP, et al.;.Chem Sci. 2021 Apr 28;12(22):7775-7785. doi: 10.1039/d1sc00330e.
- “New drugs & regimens: drug screening and improving the predictive value of preclinical models” – TTU TB, Deutsches Zentrum für Infektionsforschung (DZIF), BMBF, 2021-2025
- “Validation of the confirmed γ-glutamylspermine synthetase GlnA3 as a target for development of novel anti-tubercular drugs” - Sub project: "Relevance of the γ-glutamylspermine synthetase GlnA3 for successful growth inhibition of M. tuberculosis" (GSS-TUBTAR) – BMBF, Programm: Targetvalidierung für die pharmazeutische Wirkstoffentwicklung II, 2021-2023
- “Isolation and Characterization of M. tuberculosis-induced lipid droplets from human primary macrophages“; Leibniz Center Infection (LCI), 2018-2021
- “Discovering new therapeutic targets and drugs to combat AMR tuberculosis: proteomics characterization and drug screening of mycobacterium-infected macrophages" – Olav Thon Foundation, Norwegen (in Kooperation with T. Flo (N), M. Lerm (SE) und K. Prasad (IND), 2018-2022
- „Exploiting the methylerythritol phosphate pathway as a source of drug targets for novel anti-infectives“ - MepAnti - Marie Skodowska-Curie Innovative Training Networks, 2020-2024
- „NaPAnti: Development of natural product-based antibacterials targeting the sliding clamp DnaN“ - Bundesministerium für Bildung und Forschung, 2019-2022
- „InhibitMycoRex: Optimization of mycobacterial thioredoxin reductase inhibitors, novel lead compounds against M. tuberculosis“ – Deutsches Zentrum für Infektionsforschung, 2019-2021
- „ETBRA: European tuberculosis regimen accelerator“ – EU / Horizon 2020 (IMI), 2020-
- Host pathogen interaction
- Isolation and cultivation of primary immune cells
- murine bone marrow derived macrophages
- human monocytes
- human monocyte-derived macrophages
- human alveolar macrophages
- Infection of primary cells with strains of the M. tuberculosis (Mtb) complex (Mtb) under BSL2/BSL3 conditions in vitro
- High Content Imaging of Mtb macrophage infection (BSL3)
- Flow cytometry of mycobacteria and Mtb infected immune cells (BSL2/BSL3)
- CRISPR/CAS9 in human immune cells and murine iBMDMs
- Transfection of human macrophages
- Quantitative RT-PCR (Light Cycler)
- Seahorse XF flux analyzer
- ELISA
- Histology and immunohistochemistry
- Isolation and cultivation of primary immune cells
- TB drug discovery
- Step wise identification of novel compounds with antimycobacterial activity
- Microtiter (up to 384 well) plate-based in vitro IC50 and MIC determination (OD600, resazurin, and/or fluorescent protein signal)
- Macrophage cytotoxicity tests using real time impedance measurements (xCelligence)
- Activity tests using M. tuberculosis-infected human primary macrophages
- Activity tests against drug susceptible, resistant and MDR clinical isolates (in coop. National Reference Center) (EUCAST and MGIT format)
- Collection of widely used and uncommon pathogenic and non-pathogenic mycobacterial strains, including NTMs (M. tuberculosis H37Rv, Erdman, HN878, H37Ra; M. bovis BCG; M. avium; M. abscessus; M. haemophilum; M. chimaera, among others)
- Time kill kinetics
- In vitro generation of drug- (mono) resistant mutants
- Fluorescent Mycobacterium tuberculosis strains
- Panel of M. tuberculosis H37Rv strains constitutively expressing (via either single copy stably integrated or replicating episomal plasmids) a range of fluorescent proteins (from blue to far red).
- Genetic tools for compound mechanism of action determination and target validation
- Panel of H37Rv fluorescent reporter strains able to rapidly (2-3 days) identify diverse compound modalities, including cell wall damage, DNA damage, electron transport chain inhibition, and iron limitation (panel is routinely being expanded)
- H37Rv CRISPRi- and tetracyline-inducible gene-specific transcriptional repression and over-expression strains (current panel of ~25, including common drug targets such as MmpL3, DprE1, and RpoB (panel is also routinely being expanded).
- Step wise identification of novel compounds with antimycobacterial activity
2026
Tulu, B, Brehm, TT, van Crevel, R, Dallenga, T, DiNardo, AR, Dheda, K, Eggeling, J, Enbiale, W, Gröschel, MI, Hao, J, Kumar, V, van Laarhoven, A, Londt, R, Prosser, G, Randall, P, Reiling, N, Rybniker, J, Schaible, UE, Schurr, E, Suarez, I, Theobald, SJ, Wilkinson, RJ & Lange, C 2026, 'Host- and pathogen-related determinants of pulmonary versus extrapulmonary tuberculosis', European Respiratory Review, Jg. 35, Nr. 179. https://doi.org/10.1183/16000617.0174-2025
Walsh, DJ, Hamid, R, Giele, T, Reiling, N, Rottmann, M, Hamed, MM & Hirsch, AKH 2026, 'Design and optimization of simplified inhibitors targeting Escherichia coli and Klebsiella pneumoniae IspE', RSC medicinal chemistry. https://doi.org/10.1039/d5md00874c
Yazgili, AS, Giotopoulou, GA, Behrend, SJ, Koops, F, Neiens, V, Meul, T, Zemke, L, Reiling, N, Goldmann, T, Stathopoulos, GT & Meiners, S 2026, 'PA200 differentially regulates the proteasome and inhibits migration of NSCLC cells', JOURNAL OF CELL SCIENCE , Jg. 139, Nr. 4. https://doi.org/10.1242/jcs.264683
2025
Abdelaziz, R, Dube, M, Mann, L, Richter, A, Robaa, D, Reiling, N, Abdel-Halim, M & Imming, P 2025, 'Synthesis and Antimycobacterial Assays of Some New Ethambutol Analogs', Molecules (Basel, Switzerland), Jg. 30, Nr. 3. https://doi.org/10.3390/molecules30030600
Braun-Cornejo, M, Platteschorre, M, de Vries, V, Bravo, P, Sonawane, V, Hamed, MM, Haupenthal, J, Reiling, N, Rottmann, M, Piet, D, Maas, P, Diamanti, E & Hirsch, AKH 2025, 'Positive Charge in an Antimalarial Compound Unlocks Broad-Spectrum Antibacterial Activity', JACS Au, Jg. 5, Nr. 3, S. 1146-1156. https://doi.org/10.1021/jacsau.4c00935
Kiefer, AF, Voltz, A, Scherzer, D, Reberšek, R, Kany, AM, Prosser, G, Neuber, M, Reiling, N, Hirsch, AKH & Müller, R 2025, 'Engineering an Artificial Myxopyronin Derivative with Enhanced Metabolic Stability via Mutasynthesis', JACS Au. https://doi.org/10.1021/jacsau.5c00950
Krysenko, S, Emani, CS, Bäuerle, M, Oswald, M, Kulik, A, Meyners, C, Hillemann, D, Merker, M, Prosser, G, Wohlers, I, Hausch, F, Brötz-Oesterhelt, H, Mitulski, A, Reiling, N & Wohlleben, W 2025, 'GlnA3Mt is able to glutamylate spermine but it is not essential for the detoxification of spermine in Mycobacterium tuberculosis', JOURNAL OF BACTERIOLOGY, S. e0043924. https://doi.org/10.1128/jb.00439-24
Nitschkowski, D, Vierbuchen, T, Heine, H, Behrends, J, Reiling, N, Reck, M, Rabe, KF, Kugler, C, Ammerpohl, O, Drömann, D, Muley, T, Kriegsmann, M, Stathopoulos, GT, Arendt, KAM, Goldmann, T & Marwitz, S 2025, 'SMAD2 linker phosphorylation impacts overall survival, proliferation, TGFβ1-dependent gene expression and pluripotency-related proteins in NSCLC', BRITISH JOURNAL OF CANCER . https://doi.org/10.1038/s41416-025-02970-1
Palme, PR, Grover, S, Abdelaziz, R, Mann, L, Kany, AM, Ouologuem, L, Bartel, K, Sonnenkalb, L, Reiling, N, Hirsch, AKH, Schnappinger, D, Rubinstein, JL, Imming, P & Richter, A 2025, 'Design, Synthesis, and Biological Evaluation of Mono- and Diamino-Substituted Squaramide Derivatives as Potent Inhibitors of Mycobacterial Adenosine Triphosphate (ATP) Synthase', JOURNAL OF MEDICINAL CHEMISTRY. https://doi.org/10.1021/acs.jmedchem.5c02284
Riegler-Berket, L, Gödl, L, Polidori, N, Aschauer, P, Grininger, C, Prosser, G, Lichtenegger, J, Sagmeister, T, Parigger, L, Gruber, CC, Reiling, N & Oberer, M 2025, 'M. tuberculosis meets European lead factory – Identification and structural characterization of novel Rv0183 inhibitors using X-ray crystallography', Diseases and Therapeutics, Jg. 1. https://doi.org/10.1016/j.dist.2025.100002
2024
Braun-Cornejo, M, Ornago, C, Sonawane, V, Haupenthal, J, Kany, AM, Diamanti, E, Jézéquel, G, Reiling, N, Blankenfeldt, W & Maas, P et al. 2024, 'Target-Directed Dynamic Combinatorial Chemistry Affords Binders of Mycobacterium tuberculosis IspE', ACS omega, Jg. 9, Nr. 36, S. 38160-38168. https://doi.org/10.1021/acsomega.4c05537
Kotimoole, CN, Ramya, VK, Kaur, P, Reiling, N, Shandil, RK, Narayanan, S, Flo, TH & Prasad, TSK 2024, 'Discovery of Species-Specific Proteotypic Peptides To Establish a Spectral Library Platform for Identification of Nontuberculosis Mycobacteria from Mass Spectrometry-Based Proteomics', JOURNAL OF PROTEOME RESEARCH , Jg. 23, Nr. 3, S. 1102-1117. https://doi.org/10.1021/acs.jproteome.3c00850
Sao Emani, C & Reiling, N 2024, 'The efflux pumps Rv1877 and Rv0191 play differential roles in the protection of Mycobacterium tuberculosis against chemical stress', Frontiers in Microbiology, Jg. 15, S. 1359188. https://doi.org/10.3389/fmicb.2024.1359188
2023
Du, X, Sonawane, V, Zhang, B, Wang, C, de Ruijter, B, Dömling, ASS, Reiling, N & Groves, MR 2023, 'Inhibitors of Aspartate Transcarbamoylase inhibit Mycobacterium tuberculosis growth', ChemMedChem, S. e202300279. https://doi.org/10.1002/cmdc.202300279
Johannsen, S, Gierse, RM, Olshanova, A, Smerznak, E, Laggner, C, Eschweiler, L, Adeli, Z, Hamid, R, Alhayek, A, Reiling, N, Haupenthal, J & Hirsch, AKH 2023, 'Not Every Hit-Identification Technique Works on 1-Deoxy-d-Xylulose 5-Phosphate Synthase (DXPS): Making the Most of a Virtual Screening Campaign', ChemMedChem, Jg. 18, Nr. 11, S. e202200590. https://doi.org/10.1002/cmdc.202200590
Kotimoole, C, Antil, N, Kasaragod, S, Behera, S, Arvind, A, Reiling, N, Flo, T & Prasad, T 2023, 'Development of a spectral library for the discovery of altered genomic events in Mycobacterium avium associated with virulence using mass spectrometry-based proteogenomic analysis', MOLECULAR & CELLULAR PROTEOMICS, Jg. 22, Nr. 5, S. 100533. https://doi.org/10.1016/j.mcpro.2023.100533
Seitz, L, Reiling, N & Hilgeroth, A 2023, 'Synthesis and Evaluation of Novel Substituted N-Aryl 1,4-Dihydropyridines as An-tituberculostatic Agents', Medicinal Chemistry. https://doi.org/10.2174/1573406419666230622121512
2022
Aryal N, Chen J, Bhattarai K, Hennrich O, Handayani I, Kramer M, Straetener J, Wommer T, Berscheid A, Peter S, Reiling N, Brötz-Oesterhelt H, Geibel C, Lämmerhofer M, Mast Y, Gross H. High Plasticity of the Amicetin Biosynthetic Pathway in Streptomyces sp. SHP 22-7 Led to the Discovery of Streptcytosine P and Cytosaminomycins F and G and Facilitated the Production of 12F-Plicacetin. J NAT PROD. 2022 Mar 25;85(3):530-539. doi: 10.1021/acs.jnatprod.1c01051. Epub 2022 Mar 9.
Brandenburg J, Heyckendorf J, Marwitz F, Zehethofer N, Linnemann L, Gisch N, Karaköse H, Reimann M, Kranzer K, Kalsdorf B, Sanchez-Carballo P, Weinkauf M, Scholz V, Malm S, Homolka S, Gaede KI, Herzmann C, Schaible UE, Hölscher C, Reiling N, Schwudke D. Tuberculostearic Acid-Containing Phosphatidylinositols as Markers of Bacterial Burden in Tuberculosis. ACS INFECT DIS. 2022 Jul 8;8(7):1303-1315. doi: 10.1021/acsinfecdis.2c00075.
Gisch N, Utpatel C, Gronbach LM, Kohl TA, Schombel U, Malm S, Dobos KM, Hesser DC, Diel R, Götsch U, Gerdes S, Shuaib YA, Ntinginya NE, Khosa C, Viegas S, Kerubo G, Ali S, Al-Hajoj SA, Ndung'u PW, Rachow A, Hoelscher M, Maurer FP, Schwudke D, Niemann S, Reiling N, Homolka S. Sub-Lineage Specific Phenolic Glycolipid Patterns in the Mycobacterium tuberculosis Complex Lineage 1. FRONT MICROBIOL. 2022 Mar 8;13:832054. doi: 10.3389/fmicb.2022.832054.
Hansen J, Kolbe K, König IR, Scherließ R, Hellfritzsch M, Malm S, Müller-Loennies S, Zallet J, Hillemann D, Wiesmüller KH, Herzmann C, Brandenburg J, Reiling N. Lipobiotin-capture magnetic bead assay for isolation, enrichment and detection of Mycobacterium tuberculosis from saliva. PLOS ONE. 2022 Jul 15;17(7):e0265554. doi: 10.1371/journal.pone.0265554.
Richter A, Seidel RW, Goddard R, Eckhardt T, Lehmann C, Dörner J, Siersleben F, Sondermann T, Mann L, Patzer M, Jäger C, Reiling N, Imming P. BTZ-Derived Benzisothiazolinones with In Vitro Activity against Mycobacterium tuberculosis. ACS MED. CHEM. LETT. 2022, July 25, 2022 epub ahead of print. https://doi.org/10.1021/acsmedchemlett.2c00215
Zhu D, Johannsen S, Masini T, Simonin C, Haupenthal J, Illarionov B, Andreas A, Awale M, Gierse RM, van der Laan T, van der Vlag R, Nasti R, Poizat M, Buhler E, Reiling N, Müller R, Fischer M, Reymond JL & Hirsch A. Discovery of novel drug-like antitubercular hits targeting the MEP pathway enzyme DXPS by strategic application of ligand-based virtual screening. CHEM SCI. 2022, doi: 10.1039/D2SC02371G Epub 2022 Aug 8.
Head

Scientific staff


Technical staff